
Before considering the glucocorticoids, first use the link to the left to learn more about steroids in general.

|
MOLECULAR & CELLULAR
NEUROBIOLOGY
Master Course Cognitive Neuroscience - Radboud
University, Nijmegen
|
|
|
|
Chapter 2: Cells and within cells |
|
|
|
|
|
|
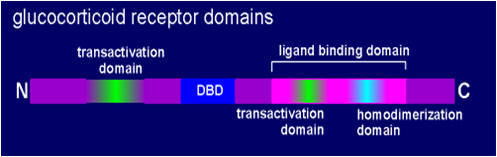
The glucocorticoid receptor is a transcription factorSimilar to all nuclear hormone receptors, the glucocorticoid receptor has a modular structure, consisting of a DNA-binding domain (DBD), a ligand-binding domain and transactivation domains to activate RNA polymerase. One region involved in transactivation is in the N terminal region of the receptor and another region with transactivation function is found within the ligand binding domain of the receptor.The DNA-binding domain is found approximately in the middle of the protein sequence. Here there are zinc fingers with 4 highly conserved cysteine molecules coordinating the binding of the zinc atom. The cysteine amino acids are indicated in yellow in the figure to the left, other amino acids in blue or purple. The formation of zinc fingers results in a structure that interacts with specific DNA sequences within the glucorticoid response element (GRE). Toward the C-terminal is the ligand binding domain. This contains a transactivation domain (already mentioned) and a homodimerization domain responsible for holding the receptor together as a dimer. The ligand binding domain is known to possess two large hyrdrphobic cavities for binding of the glucocorticoid. |
|
|
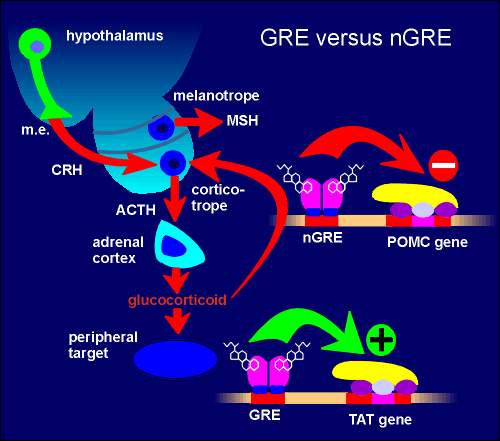
The glucocorticoid receptor can activate or repress genes The classical mode of action of the glucocorticoid receptor is via a glucocorticoid response element (GRE) on the DNA. In the absence of ligand, the receptor is trapped as an inactive cytosolic complex together with a family of proteins called heat-shock proteins. Upon ligand binding this complex dissociates and receptor dimers are formed which then translocate to the nucleus. Here it binds to the GRE. The GRE is a highly conserved sequence within the DNA. The bound receptor dimers can now interact with the basal transcriptional machinery (RNA polymerase and associated basal transcription factors). This leads to higher rates of gene transcription.
For example, the gene coding for tyrosine aminotransferase (TAT), one of the important enzymes for gluconeogenesis, has a GRE associated with it. A good example of a gene that is regulated through nGRE is the proopiomelanocortin (POMC) gene in the corticotropes of the pars distalis of the pituitary gland. In this tissue, POMC is the the precursor protein for the production of adrenocorticotropic hormone (ACTH). Through the nGRE the glucocorticoid brings about an inhibition in the production of ACTH. This is an example of negative feedback and can be regarded as a mechanism to ensure that there is not an overactivation of the hypothalamo-hypophyseal adrenal axis. Interestingly, the action of nGRE on the POMC appears to be tissue specific. For example, glucocorticoids have no effect on POMC gene expression in the melanotrope cell of the pars intermedia where the precursor protein is used to produce the peptide melanophore stimulating hormone (MSH). The ability to act through GREs or nGREs makes glucocoticoid signaling very versatile. |

|
 |
|
|
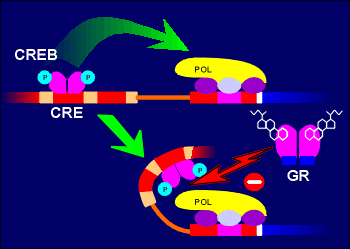 Adding
to the versatility the glucocorticoid receptor in regulatory processes has been
the finding that this receptor can also regulate gene expression by influencing
the action of other transcription factors, rather than binding directly to
responsive elements on the DNA. These DNA-binding independent mechanisms can be
positive or negative. They involve protein-protein interaction between the
activated glucocorticoid receptor and other transcription factors or cofactors
of transcription factors. For example, the glucocorticoid receptor can bind to
cAMP response element binding protein CREB (possibly through an intermediate
factor) and prevent transactivation by CREB. The net result of this action is
gene repression. Adding
to the versatility the glucocorticoid receptor in regulatory processes has been
the finding that this receptor can also regulate gene expression by influencing
the action of other transcription factors, rather than binding directly to
responsive elements on the DNA. These DNA-binding independent mechanisms can be
positive or negative. They involve protein-protein interaction between the
activated glucocorticoid receptor and other transcription factors or cofactors
of transcription factors. For example, the glucocorticoid receptor can bind to
cAMP response element binding protein CREB (possibly through an intermediate
factor) and prevent transactivation by CREB. The net result of this action is
gene repression.
|
The glucorticoid receptor acts in a similar fashion to transcription factors which act on AP-1 sites. In addition it has also been proposed that the glucocorticoid receptor inhibits cJun phosphorylation, a prerequisite for AP-1 activation. Again, the mechanism is DNA binding-independent and the net result is gene repression. Positive or synergistic effects of the glucocorticoid receptor with other transcription factors have also been reported, adding yet more versatility to glucocorticoid receptor signaling.
|
Dissecting the mechanism of glucocorticoid signaling
|
To distinguish DNA binding-dependent and
-independent mechanisms of the glucocorticoid receptor an innovative approach
has been developed.
|
|
The receptors for other steroid receptors such as for estrogen, progesterone, tesosterone and aldosterone function very similar to the glucocorticoid receptor. They too can have DNA-dependent and DNA-independent mechanisms. One (minor) difference is that some of these receptors are already in the nucleus in their inactive form (e.g. estrogen receptor) whereas other reside in the cytoplasm in their inactive form.
|
|
|
| Next page: From genome to genetic........ | Go back to: Transcription and signalling |
|
|