|
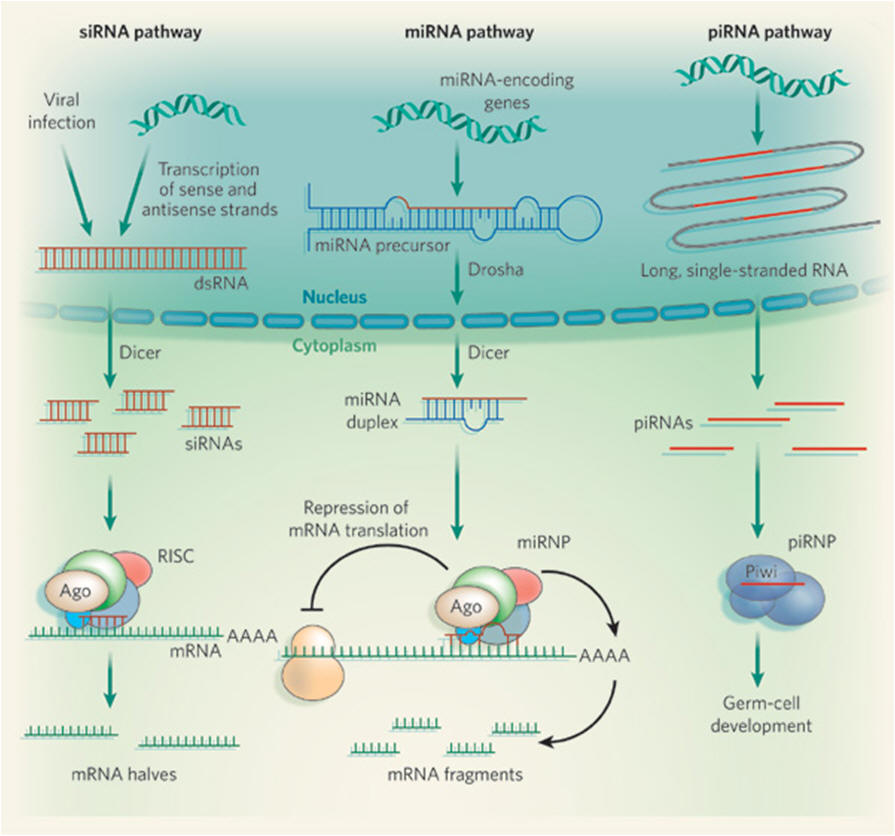
Where do the dsRNAs come from?
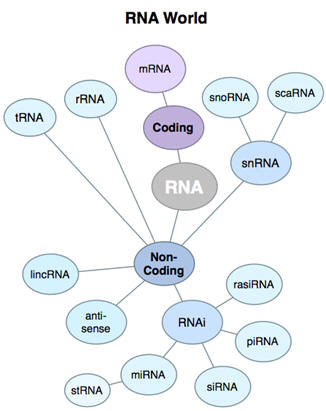
Depending on the class of small RNA, the source of precursor dsRNA differs. For
siRNAs, dsRNA can form when complementary DNA strands are transcribed into RNA
sequences. Viral infection of a cell can also supply dsRNAs, as many viruses
form RNAs of both sense and antisense polarity during replication of their
genomes, and this can trigger an RNAi response by the cell, as part of its
antiviral defence. By contrast, miRNAs are excised from 'purpose-built',
genome-encoded RNA precursors that fold into long hairpins resembling dsRNA. The
expression of miRNA-encoding genes and those encoding mRNAs are controlled very
similarly and involve the same RNA-synthetic machinery, including the enzyme RNA
polymerase II.
How do small RNAs function?
They recognize their RNA targets by sequence-specific base-pairing. The outcome
of the small-RNA–mRNA association depends on the degree of complementarity
between the two sequences. When base-pairing is perfect, or almost perfect, as
is the case for siRNAs (and possibly piRNAs), the target mRNA is cleaved in the
middle of the small-RNA–mRNA duplex. Most plant miRNAs and some animal miRNAs
function similarly. But most animal miRNAs base-pair imprecisely with mRNAs to
repress their translation or to induce their breakdown. Irrespective of
base-pairing precision, small RNAs rely on proteins of the Argonaute family for
their activity. In fact, it is the protein partners of small RNAs that bring
about repression of translation or mRNA cleavage; small RNAs act only as guides
to tell Argonaute proteins which mRNAs to target.
So can their mRNA targets be predicted through sequence analysis?
Animal miRNAs mostly base-pair to their mRNA targets with limited
complementarity. Although sequence analyses have revealed some criteria for
interaction between miRNA and mRNA, and many bioinformatics tools for target
identification are available, sequence-based predictions frequently yield false
positives or miss true targets. So identification of bona fide miRNA targets
requires extensive experimentation. By contrast, most targets of siRNAs and
plant miRNAs can be reliably predicted on the basis of near-perfect sequence
complementarity.
To prevent protein synthesis, isn't it simpler to stop mRNA production?
Intuitively, terminating transcription seems a much more obvious mechanism. But
a block in protein production always lags behind a block in transcription — even
if transcription is stopped, mRNA sequences that have already been made can
still be translated into proteins. So by targeting the existing mRNA pool, small
RNAs block or attenuate protein synthesis very rapidly and, occasionally, even
reversibly. In addition, because individual small RNAs can simultaneously target
tens if not hundreds of mRNAs for different proteins, they are well suited to
coordinate the expression of genes that function in the same or related
pathways. For example, during zebrafish embryonic development, a specific miRNA,
miR-430, targets hundreds of mRNAs for rapid degradation, facilitating
embryos' transition to a new developmental programme that requires a separate
set of proteins. The ability of different miRNAs to concurrently target several
sequences of the same mRNA further increases their potential to fine-tune gene
expression.
Is all of this
small-RNA-mediated regulation post-transcriptional?
No — small RNAs also
affect DNA transcription, particularly in plants and fission yeast. They do this
by sequence-specific targeting of chromatin (complexes of DNA with histone
proteins), converting it to the heterochromatin form that is not easily
accessible to the transcriptional machinery. Strikingly, in some lower
eukaryotes small RNAs also direct massive genomic DNA rearrangements. In
mammals, however, there is currently only limited evidence for small-RNA
functions other than post-transcriptional regulation.
Do small RNAs always silence gene expression?
In some conditions, small RNAs may also activate gene expression, although the
mechanisms are currently not well understood. Indeed, a liver-specific miRNA,
miR-122, is even needed for successful replication of the hepatitis C virus.
What biological processes do small RNAs regulate?
miRNAs were originally identified in C. elegans for their central role in
development. Consistent with their function in differentiation and development,
expression of many miRNAs is tissue-specific or is associated with certain
developmental stages. miRNA expression patterns often change in diseases such as
cancer. And, as many of the known and predicted miRNA targets have roles in
disease, it is widely believed that dysregulation of miRNA expression
contributes to disease pathology. It is less clear whether siRNAs have similarly
important functions, although in plants they have already been identified as
essential players in the regulation of stress resistance. In fission yeast and
plants, siRNAs contribute to heterochromatin formation.
And what is the link to
viruses?
The use of RNAi as a
defence mechanism against viruses may have been a driving force in the evolution
of the siRNA pathway. In plants, siRNAs are an essential layer of antiviral
defence. Also, in plants and invertebrates, siRNAs silence mobile genetic
elements (transposons), which would otherwise 'jump' around the genome and
disrupt cellular genes. It is not well known whether these small-RNA functions
are also crucial in vertebrates, in which the invention of a protein-based
adaptive immune response may have reduced reliance on antiviral RNAi activity.
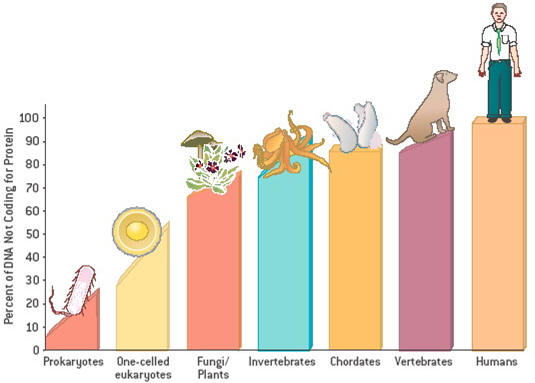
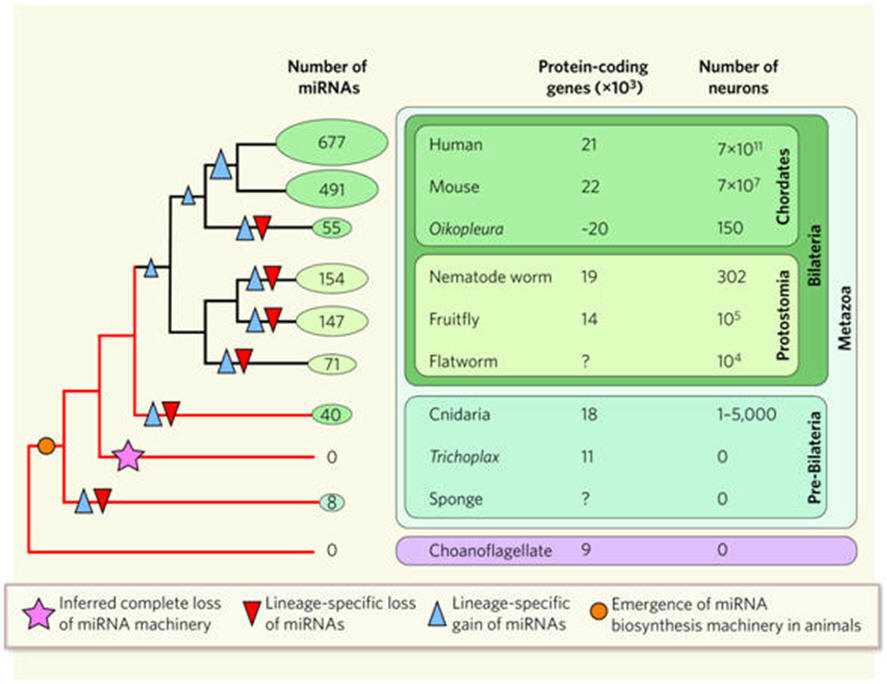
Does our collection of small RNAs set us apart from other species?
Some miRNAs are highly conserved, but others vary greatly among organisms; some
differ even among primates, for example between apes and humans. With the
emerging view that regulation of protein activity could be as vital to evolution
as fidelity of the protein sequence itself, it is tempting to speculate that
miRNAs influence evolution. Relatively simple requirements for miRNA–mRNA
interaction facilitate the development of new regulatory relationships between
these sequences, possibly contributing to the evolution of new functions (see
below; Figure 2). Perhaps, therefore, it is not surprising that a large fraction
of tissue-specific miRNAs operates in the brain.
Are small RNAs
restricted to the cells in which they are made?
In C. elegans and
plants, dsRNAs or siRNAs can move between cells or even longer distances. For
example, siRNAs spread through the vascular system of plants, which possibly
aids their function in antiviral immunity. And in C. elegans, an efficient
system of siRNA amplification ensures the maintenance of gene silencing even
after the initial 'trigger siRNA' is gone, allowing siRNA to spread to the
organism's progeny. A similar amplification system does not occur in mammals. As
for miRNAs, their very specific localization patterns, and the absence of
developmental changes in C. elegans mutants of the dsRNA transport machinery,
suggests that miRNAs are stationary.
Will small RNAs be useful as therapeutic agents or targets?
This is a hot topic of research. siRNAs have the potential to silence
disease-relevant genes that cannot be shut down with available drugs. Moreover,
the so-called oncomiRs — miRNAs that promote cancer — may themselves be targets
for shut-down. But we are still a long way from translating activity observed in
a defined experimental system into an effective therapeutic drug. One of the
most problematic issues is how to get small RNAs efficiently and specifically to
their target site of action in the human body. But regardless of their
therapeutic potential, siRNAs have already revolutionized basic biomedical
research. The use of synthetic siRNAs, or their short hairpin RNA or dsRNA
precursors, allows researchers to repress the function of a gene of interest or
even to perform genome-wide RNAi screens to unravel entire biological pathways
with unprecedented ease and speed.
So what of the future?
New classes of small RNAs continue to be discovered, and it is unlikely that we
have found them all. Even for the known classes, we often have only a very
limited understanding of what they do and how they do it. Identification of
miRNA targets is another challenge, as is identifying other, currently
hypothetical, modes of small-RNA action. Owing to their base-pairing potential,
small RNAs could modify local mRNA structures, allowing for alternative mRNA
splicing and modulating interactions of mRNAs with proteins. |