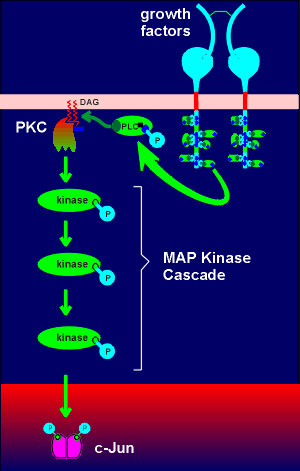
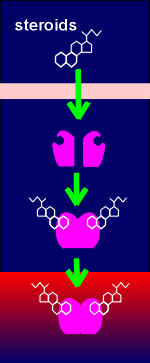
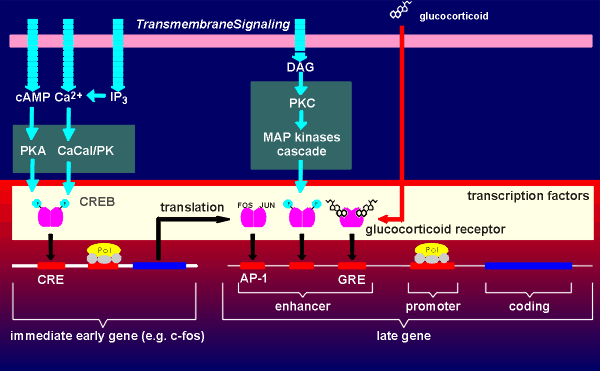
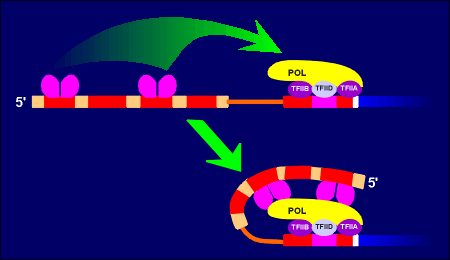
A gene has two regions, a regulatory region and a coding region. The coding region is in the 3' end of the gene (so-called downstream). The coding region is transcribed into RNAby the enzyme RNA polymerase II. The RNA produced is called the primary transcript (heteronuclear RNA). It is further enzymatically processed to give rise to the mature messenger RNA (mRNA) which is subsequently translated into protein.
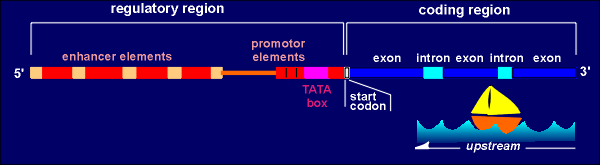
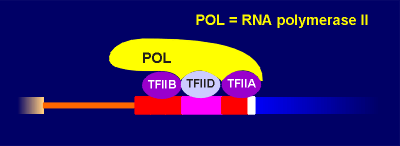
In the processing of the primary transcript the introns are cut out and the exons (ex = expression) are spliced together to form the mRNA.In the regulatory region one finds the so-called TATA box and promoter elements where transcription factors (with such names as Transcription Factor IIB etc) bind. These transcription factors, collectively called basal transcription factors, form a binding site for the RNA polymerase. They and the polymerase are sufficient to give a very low level of basal transcription of the gene into RNA.
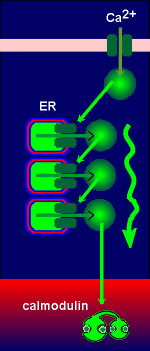
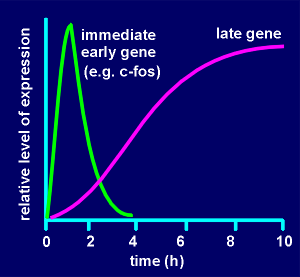
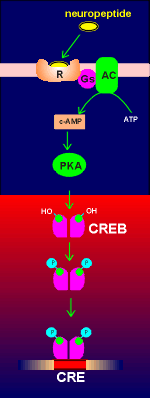 G-protein-coupled receptor mechanisms generating
cyclic AMP can lead to phosphorylation and activation of a transcription factor
called CREB (Cyclic-AMP Responsive Element Binding protein). As its name
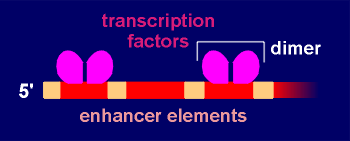
implies, CREB binds to an enhancer element on the gene called CRE (Cyclic-AMP
Responsive Element). CREB is a homodimer with phosphorylation sites for protein
kinase A (PKA). Upon activation the free catalytic subunit of PKA enters the
nucleus (nucleus indicated in red in figure to right) where it phosphorylates
CREB, which in its phosphorylated form can bind to CRE and enhance gene
expression. CREB received its name for the action of cyclic AMP on its activity.
G-protein-coupled receptor mechanisms generating
cyclic AMP can lead to phosphorylation and activation of a transcription factor
called CREB (Cyclic-AMP Responsive Element Binding protein). As its name
implies, CREB binds to an enhancer element on the gene called CRE (Cyclic-AMP
Responsive Element). CREB is a homodimer with phosphorylation sites for protein
kinase A (PKA). Upon activation the free catalytic subunit of PKA enters the
nucleus (nucleus indicated in red in figure to right) where it phosphorylates
CREB, which in its phosphorylated form can bind to CRE and enhance gene
expression. CREB received its name for the action of cyclic AMP on its activity.