 There are a
number of CREB proteins.
There are a
number of CREB proteins. There are a
number of CREB proteins.
There are a
number of CREB proteins.
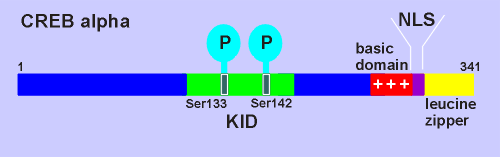
The domain structure of a typical CREB protein, CREB alpha, is shown on the left.
This is a protein of 341 amino acids.
Approximately in the middle of the protein is the kinase inducible domain (KID) with two sites for Ser/Thr kinase phosphorylations, namely Ser133 and Ser142.
Phosphorolation at Ser133 has proved to be the critical event for CREB activation.
 At the extreme C
terminal is the leucine zipper domain responsible for protein-protein interaction to form
dimers (leucine zippers were also used to form dimers in the PKG and PKC; review kinase structure?).
At the extreme C
terminal is the leucine zipper domain responsible for protein-protein interaction to form
dimers (leucine zippers were also used to form dimers in the PKG and PKC; review kinase structure?).
Also in the C terminal region is the nuclear localization signal (NLS), a small sequence which directs CREB to the nucleus following its biosynthesis.
Most CREB proteins are found in the nucleus.
DNA binding of CREB to CRE (cyclic AMP responsive element) is mediated by the basic domain, a lysine- and arginine-rich stretch of amino acids in the C-terminal region.
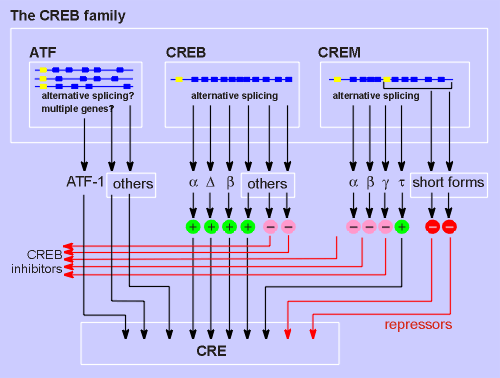 As mentioned, CREB is
not a single protein but rather is a large family of proteins.
As mentioned, CREB is
not a single protein but rather is a large family of proteins.
First there is the CREB gene itself which through alternative splicing produces a number of different CREB isoforms such as alpha, delta and beta.
The exons in the CREB gene are indicated by the think blue line; the CREB gene has 11 exons.
Most CREBs act as activating transcription factors (green circles) but some isoforms act as inhibitors (pink circles).
Possibily these inhibitory isoforms bind CRE, thus blocking the binding of activator forms of CREB.
Alternatively, they may dimerize with the activator forms to produce an inactive dimer.
A highly related gene family is the activating transcription factors (ATF) family (members of which are thought to be generated by multiple genes or alternative splicing).
One of these, ATF-1 is certainly in the CREB family, with 65% identical to CREB in primary structure.
Other ATFs may or may not be in the CREB family.
The third gene family within the CREB family is CREM, cyclic AMP response element modulator.
Again, this is a single gene with alternative splicing generating many different isoforms of the protein.
Many of the CREMs are CREB inhibitors (e.g. the alpha, beta and gamma form), possibily blocking the CRE site or dimerizing with CREB to produce an inactive dimer.
One, called CREM(tau), acts as an activator of CRE-mediated transcription similar to CREB itself.
Two particularly short forms of CREM are generated through the use of either internal translation initiation or internal intronic promoter (promoters indicated in yellow in above diagram).
These two isoforms act as CRE-binding repressors (i.e. they bind CRE and inhibit the transcription machinery itself rather than just blocking CREB activity, as do the inhibitors).
CREB, ATF-1 and CREM are able to bind CRE as homodimers, but they also are know to form heterodimers (not surprising as they have highly related dimerization domains).
Which ATF-1,CREB, CREM combinations actually form in living cells, and the exact nature of the activities of such hetrodimers, remains to be determined.
It is clear however that the CREB family of proteins has great potential to generate a large variety of signaling possibilities.
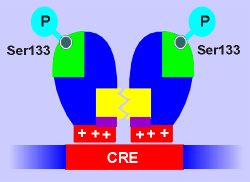
Stimulus-induced activation of CREB is mediated by phosphorylation.
How does phosphorylation of CREB stimulate its ability to activate transcription?
One idea is that phosphorylation may induce dimerization (a prerequisite for gene activation) but there is no compelling evidence for this.
Another idea is that phosphorylation of CREB may increase its affinity for CRE but, again, compelling evidence is lacking.
This led researchers to search for factors which might associate with CREB in a phosphorylation-dependent manner.
This search invloved looking for proteins which would bind radioactive CREB.
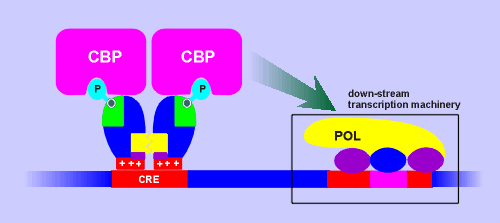 A protein was isolated which specifically
bound Ser133-phosphorylated CREB.
A protein was isolated which specifically
bound Ser133-phosphorylated CREB.
The protein was given the name CREB-binding protein (CBP).
Data quickly accumulated showing that CBP was critical for the functioning of CREB.
For example, microinjecting cells with antibodies against CBP inhibited cyclic AMP-induced activation of genes.
CBP has proved to be a transcriptional adaptor protein which bind phophorylated CREB on the CRE site and subsequently interacts with and activates the down-stream basal transcription machinery.
Action through adaptor proteins is very common among transcription factors.
When first discovered, CRE was thought to be a responsive element strictly for the cyclic AMP system, hence the name CRE and CREB.
This has turned out to be completely wrong.
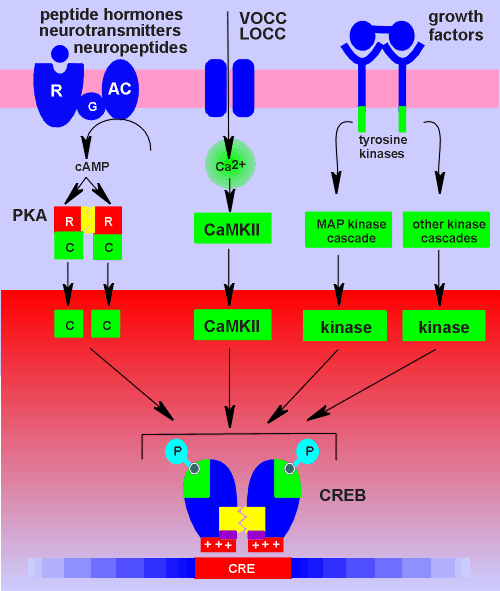 In fact, CREB can be
considered an intergrating machine because it is the focal point for a number of
converging kinase pathway.
In fact, CREB can be
considered an intergrating machine because it is the focal point for a number of
converging kinase pathway.
The figure to the left illustrates some of the pathways converging on CREB.
All three catagories of transmembrane signaling can activate CREB.
For G protein-coupled receptors the route is obvious, via cyclic AMP and PKA.
A ligand-operated Ca2+ channel (LOCC), such as the NMDA receptor, can activate CREB via Ca2+ calmodulin-dependent protein kinase II (CaMkII).
This kinase also phosphorylates Ser133 in CREB.
The source of the Ca2+ could equally come from a voltage-operated Ca2+ channel (VOCC).
Likewise, the source of the Ca2+ could be intracellular, such as from activation of IP3 receptors (this possibility is not shown in figure).
In this way neuropeptides and peptide hormones working via Gq can signal via CREB.
Finally, growth factors, such as nerve growth factor or brain-derived neurotropic factor (BDNF), working through tyrosine kinase receptors can feed in on CREB.
They do this through activation of a series of kinases, known as the MAP kinase cascade, which ultimately leads to phosphoryaltion of CREB.
It has also become clear that regulation of CREB activity involves more than just phosphorylation at Ser133.
Other phosphorylation events on CREB appear to regulate the ability of CREB to induce expression of specific target genes in response to particular stimuli.
The convergence of many kinase system on CREB Ser133 allows CREB to function as a barometer of envronmental change; at the same time the requirement for other phosphorylation events may allow CREB to adjust specific cellular responses to meet specific stimuli.