 The gene is
regulated by CREB and another transcription factor call serum response factor
(SRF) binding to serum response element (SRE).
The gene is
regulated by CREB and another transcription factor call serum response factor
(SRF) binding to serum response element (SRE).
This is a beautiful example of an innovative approach in research leading to an important new finding.
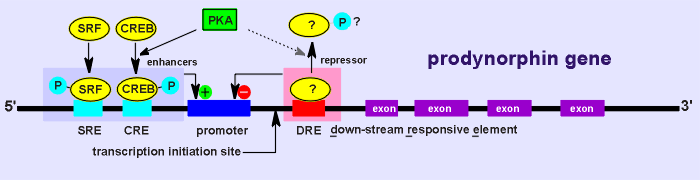
The story concerns the regulation of the prodynorphin gene (dynorphin is a neuropeptide of the opiate family).
Given below is the dynorphin gene structure.
 The gene is
regulated by CREB and another transcription factor call serum response factor
(SRF) binding to serum response element (SRE).
The gene is
regulated by CREB and another transcription factor call serum response factor
(SRF) binding to serum response element (SRE).
As well, from the gene structure it was known that the gene had a down-stream responsive element, DRE.
There was some evidence that this element was the binding site for a repressor (a transcription factor which represses gene expression).
It was even hypothesized that a phosphorylation (perhaps through PKA) might regulate this factor (as depicted in the diagram above).
 A very innovative
approach was developed to go after the repressor.
A very innovative
approach was developed to go after the repressor.
It was hypothesized that the repressor, whatever it was, should be expressed in a human brain expression library.
This library was constructed by extracting mRNA from the human brain, making cDNA using the mRNA as template, and then putting the cDNA into an expression vector (a vector which, in the bacteria would express the cDNA as protein).
What was lacking was a probe to pull the repressor out of the library (i.e. to identify which bacterial clones are expressing the repressor).
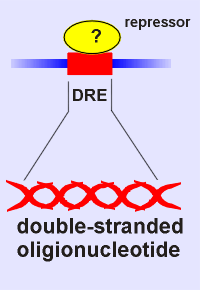 What was known was that
the repressor would bind DRE.
What was known was that
the repressor would bind DRE.
Thus came the idea, make a probe of DRE!
So a radioactive double-stranded oligionucleotide was produced with the DRE nucleotide sequence.
 This was then used to probe
the expression library to see if there were any clones producing a protein that would bind
specifically the DRE probe.
This was then used to probe
the expression library to see if there were any clones producing a protein that would bind
specifically the DRE probe.
A colony that bound the probe was indeed found.
The cDNA was extracted out of this colony, amplified and then sequenced.
It was found that it coded for a 32 K protein, and further test showed that it acted as a repressor of the prodynorphin gene.
It was named DRE-antagonist modulator or DREAM.
[This is a rather unfortunate designation as it is neither an antagonist nor a modulator; perhaps DREB (Downstream Responsive Element Binding protein, in analogy with the naming of CREB, did not sound flashy enough, particularly for a Nature article!]
 Once they had the
protein (from the expression library) they could examine its properties (i.e. make an
antibody to check out its localization; examine if it indeed is a substrate for PKA).
Once they had the
protein (from the expression library) they could examine its properties (i.e. make an
antibody to check out its localization; examine if it indeed is a substrate for PKA).
DREAM proved to have some interesting properties (see list to left).
As expected it was highly expressed in the brain and located in the nucleus.
It was not a PKA substrate! Thus the earlier idea that phosphorylation of this repressor would regulate its activity proved wrong.
DREAM possessed 4 potential Ca2+ binding domains and it was shown that DREAM indeed could bind Ca2+.
Moreover it was shown that the Ca2+ binding blocked DREAM binding to the CRE probe.
Thus it was hypothesized that DREAM is a Ca2+ sensor in the nucleus involved in gene regulation.
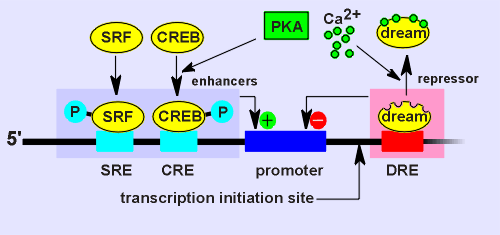 The idea is that
Ca2+ would regulate the activity of the repressor.
The idea is that
Ca2+ would regulate the activity of the repressor.
Under low Ca2+ conditions DREAM would bind DRE and thus repress the gene.
Under high Ca2+ conditions (such as a Ca2+ wave entering the nucleus) the Ca2+ would bind to DREAM and DREAM would leave the DRE site (see diagram to right).
Consequently the gene repression would be lifted.
This was the theory.
The question was, would it really work this way in a living cell?
Experiments were designed to test if indeed DREAM acts as a Ca2+ sensor in the regulation of gene activity.
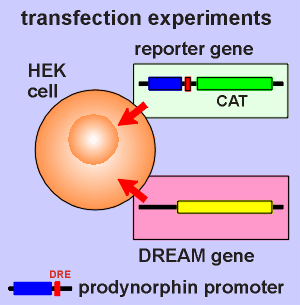 For this purpose the
prodynorphin promoter region (including DRE) was fused to DNA coding for a product
that is easily measured.
For this purpose the
prodynorphin promoter region (including DRE) was fused to DNA coding for a product
that is easily measured.
In this case it was the bacterial enzyme chloramphenicol acetyltransferase (CAT), the activity of which can be easily measured. (commercial kits are available to do this).
This is called the reporter gene (in this case it will report the expression level of the dynorphin promoter).
The reporter gene construct was transfected into cultured tumor cells called HEK cells (human embryonic kidney cells).
The DREAM gene was also transfected into the same cells and tests were conducted to confirm that the transfected cells expressed DREAM.
So in the HEK cell we have DREAM, and we have a promoter sensitive to DREAM driving CAT expression. We also have a CAT kit.
The next step is to see if Ca2+ has an effect on the expression of CAT.
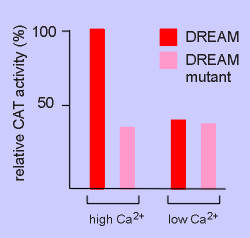 The results are
shown to the right.
The results are
shown to the right.
In the cell transfected with DREAM the expression of CAT was high under high Ca2+ conditions and low under low Ca2+ conditions.
Thus the hypothesis seems correct.
But is this action of Ca2+ really coming through action on DREAM (and not, for example through Ca2+ induced phosphorylation of CREB, that might also be found in HEK cells).
A nice control experiment was included to check this out.
A mutated form of DREAM was made which could still bind to DRE (and thus supress gene activity) but could no longer bind Ca2+ (mutations in the Ca2+ binding domains).
Cells transfected with the Ca2+ insensitive DREAM mutant displayed low CAT activity and the high Ca2+ failed to overcome this action.
Thus it would appear that DREAM itself is indeed acting as a Ca2+ sensor in the regulation of gene expression!
DREAM is the first functional nuclear Ca2+ sensor (there are other Ca2+ binding proteins in the nucleus, such as calmodulin, but the function of these are less well established).