Genomic and cDNA libraries
Genomic and cDNA libraries are made to identify a gene and a cDNA, respectively.
Construction of a genomic library
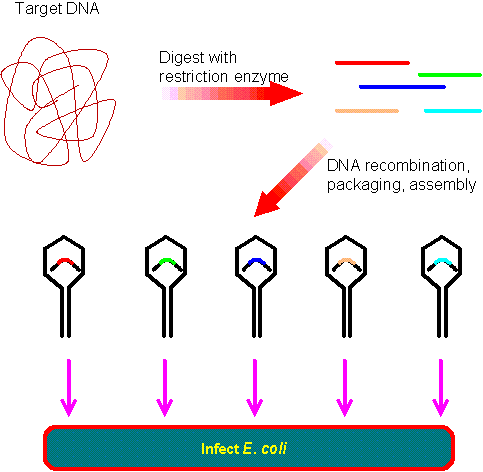
A genomic library comprises a set of bacteria, each carrying a different small fragment of human DNA. For simplicity, cloning of just a few representative fragments is shown. In reality, all the DNA fragments will be cloned. The genomic library is normally made by l phage vectors (lower figure), instead of plasmid vectors (figure on the right). The genomic library contains DNA fragments representing the entire genome of an organism. The entire human genome is about 3 x 109 base pairs (bp) long while a plasmid or phage vector may carry up to 20 kb fragment. This would require 1.5 x 105 recombinant plasmids or l phages. When plating E. coli colonies on a 3" petri dish, the maximum number to allow isolation of individual colonies is about 200 colonies per dish. Thus, at least 700 petri dishes are required to construct a human genomic library. By contrast, as many as 5 x 104 l phage plagues can be screened on a typical petri dish. This requires only 30 petri dishes to construct a human genomic library. Another advantage of l phage vector is that its transformation efficiency is about 1000 times higher than the plasmid vector.
Construction of a cDNA library
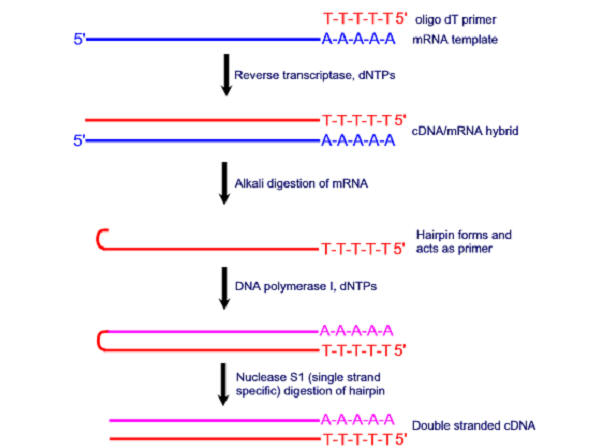
The cDNA library contains only complementary DNA (cDNA) molecules synthesized from mRNA molecules in a cell. The advantage of a cDNA library is that it contains only the coding region of a genome. To prepare a cDNA library, the first step is to isolate the total mRNA from the cell type of interest. Because eukaryotic mRNAs consist of a poly-A tail, they can easily be separated. Then the enzyme reverse transcriptase is used to synthesize a DNA strand complementary to each mRNA molecule. After the single-stranded DNA molecules are converted into double-stranded DNA molecules by DNA polymerase, they are inserted into vectors and cloned.
The cloned cDNA is then transformed into bacteria and the collection of bacteria is the cDNA library. When the cDNA library is plated on agar, individual bacterial colonies can be picked by a robotic colony picker (see Figure below).
|
|
 |
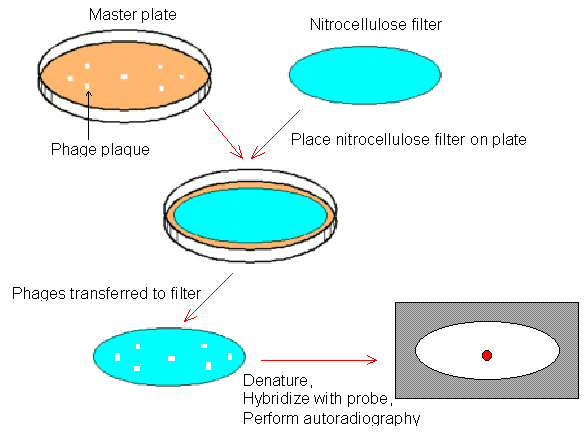
Screening a library
Screening a library is searching for a specific DNA fragment in the library. After recombinant l virions form plaques on the lawn of E. coli, the nitrocellulose filter (membrane) is placed on the surface of the petri dish to pick up l phages from each plaque. Then, the filter is incubated in an alkaline solution to disrupt the virions and release the encapsulated DNA, which is subsequently denatured. Next, the probe is added to hybridize with the target DNA fragment; the probe is a collection of single-stranded DNA or RNA molecules of specific base sequence, labeled either radioactively or immunologically, that is used to detect the complementary base sequence by hybridization; hybridization is the process of joining two complementary strands of DNA or one each of DNA and RNA to form a double-stranded molecule. The position of the probe-labelled target DNA may be displayed by autoradiography. Once a particular DNA fragment is identified, it can be isolated and amplified to determine its sequence. If we know the partial sequence of a gene and want to determine its entire sequence, the probe should contain the known sequence so that the detected DNA fragment may contain the gene of interest.