 Nicotinic receptors are found in neuromuscular junctions where motor neurons
release acetylcholine to control muscle contractions.
Nicotinic receptors are found in neuromuscular junctions where motor neurons
release acetylcholine to control muscle contractions.The nicotinic receptor is an ion channel receptor with, as endogenous ligand, the neurotransmitter acetylcholine.
The drug nicotine is also a ligand for the receptor....thus the name nicotinic receptor (there is also another receptor for acetylcholine, the acetylcholine muscarinic receptor which is a G-protein coupled receptor.......but that is another story).
 Nicotinic receptors are found in neuromuscular junctions where motor neurons
release acetylcholine to control muscle contractions.
Nicotinic receptors are found in neuromuscular junctions where motor neurons
release acetylcholine to control muscle contractions.
These receptors are also found in the brain where they are the targets for the addictive activity of nicotine.
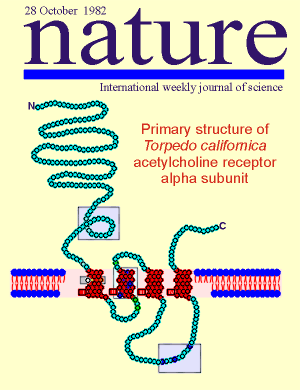 The
acetylcholine nicotinic receptor has a long history in receptor research. A high-light in
this history came in 1982.
The
acetylcholine nicotinic receptor has a long history in receptor research. A high-light in
this history came in 1982.
In that year the nicotinic receptor became the first membrane receptor molecule to which recombinant cDNA technology was applied.
Consequently, it became the first membrane receptor molecule where the full amino acid sequence of the receptor was known.
 For ~20
years prior to this biochemists had been busy with the "classical" biochemical
approach to determine the sequence of the nicotinic receptor.
For ~20
years prior to this biochemists had been busy with the "classical" biochemical
approach to determine the sequence of the nicotinic receptor.
The classical approach involves the extraction of protein from tissue possessing the receptor, purification of the receptor protein and, finally, amino acid sequencing.
One of the favorite tissues to use as a starting material by biochemists was the electric organ of the ray Torpedo californica (shown below).
The electric organ consists of plate-like cells derived from muscle tissue.
 This
organ generates pulsed electrical discharges of up to 200 volts for about 1 second.
This
organ generates pulsed electrical discharges of up to 200 volts for about 1 second.
The shocks enables torpedoes to stun faster-swimming fishes and also serves as a defense against predators.
The organ is regulated by acetylcholine working through the nicotinic receptor (the same as muscle tissue).
The tissue is relatively rich in nicotinic receptors and thus made a good starting point in studies attempting to purify and sequence the receptor.
This approach never delivered the full sequence, however, because the job was simply too big and too difficult (receptors are present in only very low amounts, there is tremendous loss of receptor protein during purification and large amounts of purified protein are required for amino acid sequencing).
As outlined below, the approach did, however, make major contributions to our understanding of the structure and functioning of the nicotinic receptor.
The classical biochemists discovered that the nicotinic receptor was constructed from subunits.
Each receptor was composed of 5 subunits that were named a, b, g and d; the a subunit was present twice and thus the receptor was referred to a2bgd.
They found that the subunits were very similar in size and structure.
Studies with radiolabeled acetylcholine showed that it bound to the a subunit; thus the nicotinic receptor has two binding sites for the endogenous ligand.
The biochemists applied "nearest-neighbor" analysis to the receptor.
This is a technique whereby isolated, intact receptor is treated with a cross-linking agent so that subunits next to each other will be covalently bound. They can subsequently determine which subunits are bound to each other and thus which subunits sit next to each other in the native structure.
The results...both a subunit bound to the g ; one of the a subunits bound to the b, the other to the d; the b and d subunits were also bound to each other.
Put it together and what have you
got?..........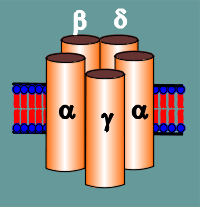
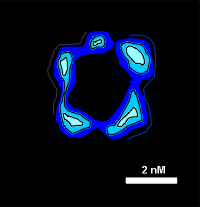
Evidence that the subunits indeed form a ring was found in electron microscopic studies of negatively stained membranes from Torpedo electric organ tissue (image to right). The computer-aided image clearly shows the presence of 5 units forming a ring-like structure.
 Enter
Dr. Numa..............................
Enter
Dr. Numa..............................Dr. Shosaku Numa ushered in a whole new era in receptor research with the recombinant DNA approach.
He had earlier ushered in a new era of precursor protein research with the application of this same approach to obtain the amino acid sequence of precurors for peptide hormones and neuropeptides (use link to right for more detail).
In the receptor field, he was able, in two years of research, to achieve something that the classical biochemists were unable to achieve in 20 years....full sequence information for the receptor.
Why was he able to achieve this in so short a time? The answer can be found if one compare the "classical biochemical" versus the recombinant DNA approach.
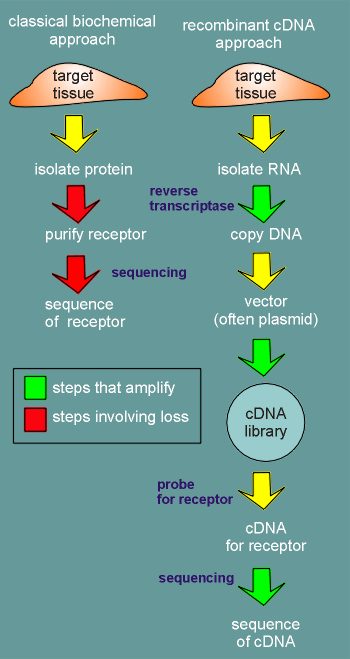 first
consider the approach used by the classical biochemist .....
first
consider the approach used by the classical biochemist .....
Receptors are present in very low amounts in tissue and they are membrane bound which makes them very difficult to isolate.
Once the membrane bound proteins have been isolated from the target tissue then the laborious job of purifying the receptor begins (protein sequencing requires pure protein).
This involves many steps using different isolation methods such as column chromatography and electrophoresis.
Losses of protein are encountered at each step.
Finally the sequencing begins. This is a destructive method where the protein is broken into small fragments for sequencing.
Again a heavy loss of material.
Now, consider the recombinant approach....
First, RNA is isolated from the target tissue.
The isolated RNA is transcribed into cDNA, an enzymatic reaction using the RNA as template.
Each RNA may be copied 10,000 times or 100,000 times...a tremendous amplification!
The cDNA is then put into a so-called vector and a cDNA library is produced.
The vector is often a plasmid that is carried by a bacteria.
Thus the cDNA library is carried by a mixture of different bacteria, each bacteria carrying a different cDNA, depending on which plasmid they are carrying.
The bacteria are plated out on agar and thereafter, each time the bacteria divides within a colony there is an amplification of the amount of cDNA (the plasmids duplicate each time the bacteria duplicate).
So we again have amplification!
 The colony on the agar plate carrying the cDNA for the
receptor is identified by "probing " for it with a probe that makes a specific
binding to the cDNA for the receptor (use the link to find out more about the probe).
The colony on the agar plate carrying the cDNA for the
receptor is identified by "probing " for it with a probe that makes a specific
binding to the cDNA for the receptor (use the link to find out more about the probe).
This colony is then selected off the plate and cultured (again involving amplification) and finally the plasmids are isolated and the cDNA cut out and sequenced.
 In cDNA
sequencing there is also an amplification of signal (in the sequencing the cDNA is used as
template to produce radiolabeled or fluorescent copies).
In cDNA
sequencing there is also an amplification of signal (in the sequencing the cDNA is used as
template to produce radiolabeled or fluorescent copies).
With the cDNA sequence one can then read-off the sequence of amino acids which would be found in the receptor.
 In
conclusion, in the classical biochemical approach there is a loss of material or
signal at a number of steps in the process.
In
conclusion, in the classical biochemical approach there is a loss of material or
signal at a number of steps in the process.
With the molecular recombinant approach there is amplification at a number of steps in the process.
It is no wonder that the classical approach has never been able to deliver the full sequence of a receptor whereas the recombinant DNA approach has been highly successful in doing so.
The next two steps in the analysis of a receptor with the recombinant approach are (1) make a hydropathy profile of the receptor and (2), with the aid of this profile examine the structure for functional domains.
First, we will look at what a hydropathy profile is.
In a hydropathy profile of a protein (such as a receptor protein) each amino acid within the protein is assigned a number to reflect its relative hydrophobicity.
In the example about to be considered, highly hydrophobic amino acids (such as tyrosine, phenylalanine) have a number near "3" whereas highly hydrophilic amino acids (such as charged amino acids like aspartic acid, glutamic acid, lysine and arginine) have a number assigned near "-3".
Neutral amino acids (such as glycine) are assigned a value of about "0".
The hydropathy profile of a protein is a plot of the relative hydrophobicity of each amino acid in the protein versus its position within the protein.
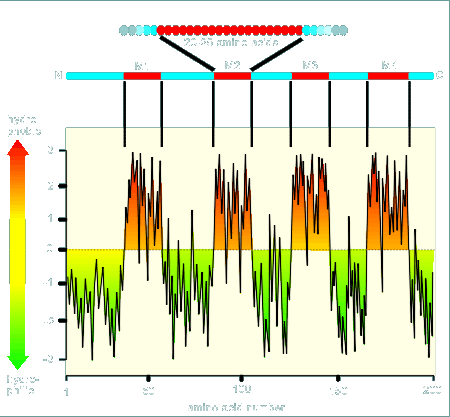 The plot starts from the
N-terminal and works its way to the C-terminal (see example at left for a 200 amino acid
protein).
The plot starts from the
N-terminal and works its way to the C-terminal (see example at left for a 200 amino acid
protein).
Notice that in the beginning there are many charged amino acids and thus the hydropathy plot is for the most part below zero.
At a certain point the protein has many hydrophobic amino acids.
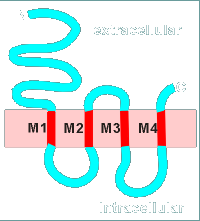
If we are dealing with a membrane bound protein and if this hydrophobic region is in a stretch of about 25 amino acids then there is a very good chance that this is a region where the protein would go over the hydrophobic membrane.
In the hydropathy plot shown there are four such regions, designated M1, M2, M3 and M4.
The prediction would be that this protein would go across the membrane four times.
Why hydrophobic amino acids in transmembrane regions? Because they are stable in the hydrophobic environment of the lipid bilayer.
Why so many amino acids to get across the lipid bilayer (one would have expected 5 or 6 amino acids to be sufficient to span the bilayer)?
 In general proteins that go across membranes go across in an alpha-helix.
This means it takes about 25 amino acids to get across the membrane.
In general proteins that go across membranes go across in an alpha-helix.
This means it takes about 25 amino acids to get across the membrane.
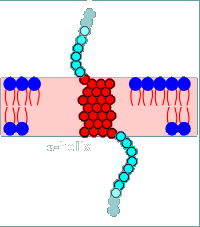
With the hydropathy profile completed one can now make a drawing (model) of how the protein will sit in the membrane.
This has been done to the left for the example protein.
Notice that the N-terminal is extracellular (this is true of all plasma membrane receptor proteins).
The protein threads itself in and out of the membrane four times corresponding to the 4 hydrophobic regions of the hydropathy profile (M1, M2, M3 and M4).
In fact this example is based on the subunits of the nicotinic receptor.
Each subunit had four hydrophobic regions as potential transmembrane regions.
Thus it has been predicted that each subunit would have the basic structure shown above.
Next, we will examine for functional domains.....
Functional domains are regions (or domains) in the receptor that are responsible for different functions the receptor must fulfill.
For example, for the nicotinic receptor we would expect at least the following domains:
1. ligand binding domain
2. ion pore domain
The hydropathy profile shows you what parts of the protein are extracellular, what parts are in the membrane and what parts are intracellular.
Thus one knows where to look for a ligand binding domain (in the extracellular region) and where, perhaps, to look for the ion pore (transmembrane region?).
Thus you look in these regions for special structures that might fulfill these functions (this is why a hydropathy profile is made first, before looking for functional domains).
As outlined in the next section, this has been done this for the nicotinic receptor to identify potential functional domains.
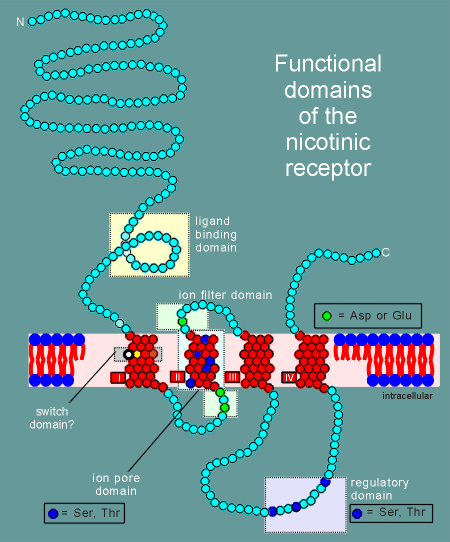 Given
to the right is a model for the alpha subunit of the nicotinic receptor, based on the
hyropathy profile and an examination of the structure.
Given
to the right is a model for the alpha subunit of the nicotinic receptor, based on the
hyropathy profile and an examination of the structure.
It includes 5 functional or potential functional domains as indicated in the figure and listed in the menu below.
Click on the various domains in the menu below to learn more about each domain.
| ligand binding domain |
| ion pore domain |
| ion filter domain |
| switch domain |
| regulatory domain |
note: The diagram gives the functional domains for the alpha subunit structure. The structure of the beta, gamma and delta subunits are very similar to this structure.
The other subunits had all the functional domains possessed by the alpha, with the exception of the ligand binding domain (remember, acetylcholine binds to the alpha subunit).
It is all very nice to look for functional domains and make models for how the receptor might work...but the important question remains.......is it all true?
Not only have the molecular biologists developed the models based on hydropathy profiles and functional domains, they have also come up with a beautiful way to test the models.
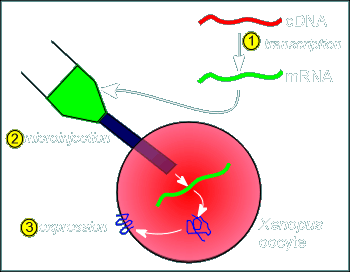 The
method involves first, taking the cDNA coding for the receptor (or the receptor subunits)
and using the enzyme transcriptase make a RNA copy (step 1 in figure at right).
The
method involves first, taking the cDNA coding for the receptor (or the receptor subunits)
and using the enzyme transcriptase make a RNA copy (step 1 in figure at right).
This RNA is then introduced into an expression system (a cell that will translate the RNA into protein).
This expression system is often oocytes of Xenopus laevis.
Why Xenopus oocytes?.....because (1) the oocyte is big (thus making injection of the RNA construct easy) and (2) the oocyte is packed full of ribosomes and all the other hardware required for protein production...once a RNA is injected it is immediately translated.
In the case of the nicotinic receptor the cDNA for the alpha, beta gamma and delta subunits are transcribed into RNA and these are injected into the oocyte (twice as much RNA coding for the alpha is injected so that we get about twice the amount of expression of the alpha subunit).
The oocyte expresses the RNA and constructs the receptor on the membrane (the RNA contains all the information for where the protein should be produced i.e. it has a coding for a signal sequence and thus synthesis occurs in the rough endoplasmic reticulum and the resulting protein finds its way to the membrane via the constitutive pathway).
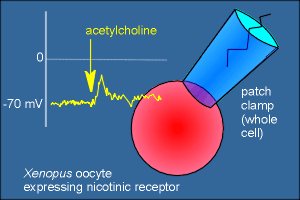 Thus, with the injection of the appropriate RNA the oocyte expresses the
nicotinic receptor and will actually depolarize if the cell is given acetylcholine or
nicotine!
Thus, with the injection of the appropriate RNA the oocyte expresses the
nicotinic receptor and will actually depolarize if the cell is given acetylcholine or
nicotine!
 This can be seen using the patch-clamp method (if you're not
familiar with this method use the link to the right to get a quick overview of the
method).
This can be seen using the patch-clamp method (if you're not
familiar with this method use the link to the right to get a quick overview of the
method).
....and now the real trick to test the model, site directed mutagenesis!
To test the model molecular biologists can, as the first step, alter the nucleotide coding within the cDNA so they can alter which amino acid a codon within the cDNA codes for.
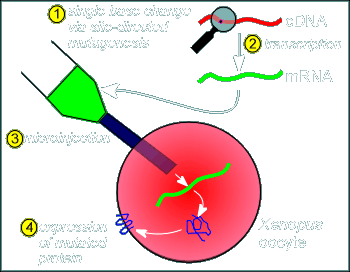 This method is called "site directed mutagenesis" (technical
details on how site directed mutations are introduced into cDNA is beyond the scope of
this tutorial; the developement of this method lead to the Nobel Prize in 1993).
This method is called "site directed mutagenesis" (technical
details on how site directed mutations are introduced into cDNA is beyond the scope of
this tutorial; the developement of this method lead to the Nobel Prize in 1993).
If the cDNA containing the mutated codon is now copied into RNA and the RNA injected into the Xenopus oocyte, as described above, then we will have expression of an altered or "mutated" protein.
This method can be used to test the model.
For example, if one wanted to examine if the aspartic acid and asparagine in the beta-structural loop are really involved in ligand binding then one would conduct site directed mutagenesis and change the coding for these amino acids in the cDNA to a coding for a neutral amino acid....then make RNA....then inject...then test the resulting receptor for its ability to depolarize with acetylcholine treatment or to bind radioactive acetylcholine.
....and, how has our model of the nicotinic receptor held up when tested in this (and other) ways?
Check it out with the links below:

As already mentioned, the biochemists had discovered that the alpha, beta, gamma and delta subunits were all about the same size.
The recombinant approach gave full sequence information for the subunits and revealed that they had a high degree of sequence homology.
This has lead to the idea that they were all derived originally from the same gene.
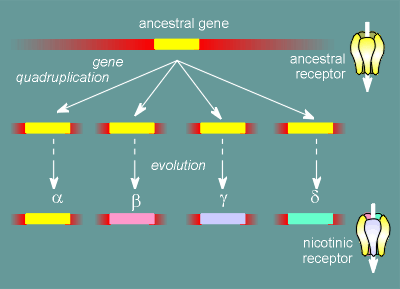 The gene underwent a quadruplication and, during evolution, these genes
mutated thus giving rise today to four different, but highly similar, proteins.
The gene underwent a quadruplication and, during evolution, these genes
mutated thus giving rise today to four different, but highly similar, proteins.
In fact this scheme is a gross over simplification.
There is a whole family of alpha, beta, gamma and delta subunits.
This means that there is a whole family of nicotinic receptors.
Which nicotinic receptor a cell possess depends on exactly which genes the cell expresses for the various subunits.
The nicotinic receptors with different subunit compositions have different properties.
For example, the nicotinic receptor in the brain is particularly sensitive to nicotine whereas the nicotinic receptor of the neuromuscular junction is rather insensitive to nicotine.
Thus, nicotine has very little effect on the function of the somatic nervous system but has addictive effects in the central nervous system.
 This
is the end of the module "The Nicotinic Receptor"
This
is the end of the module "The Nicotinic Receptor"