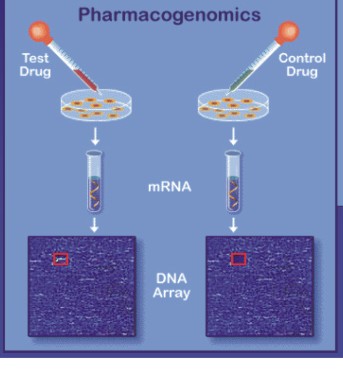
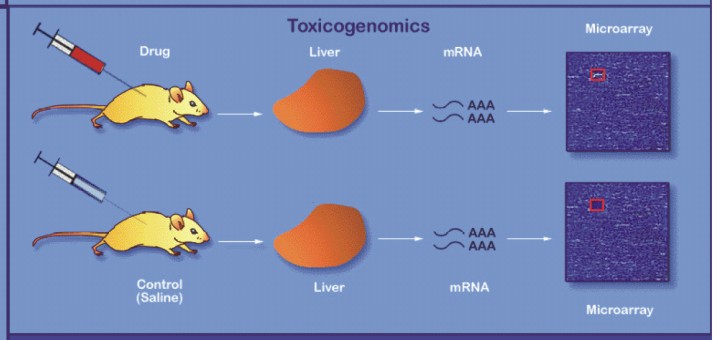
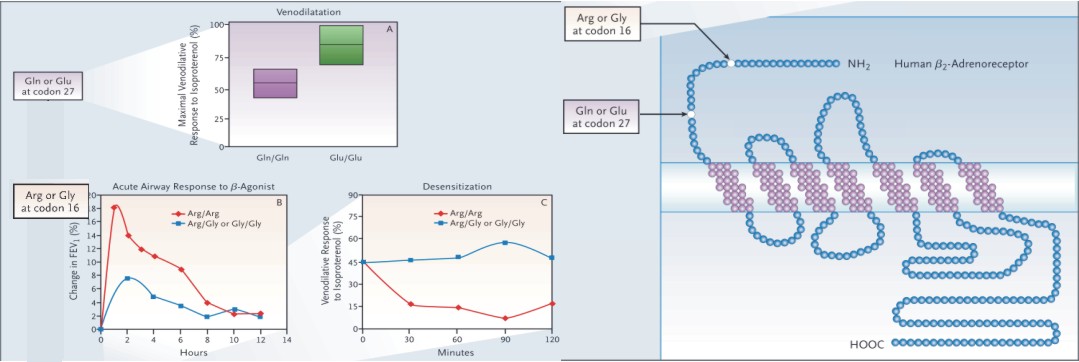
|
This field of study was called Pharmacogenetics in the late
1950s, but more recent developments expanding the breadth
and depth of the field have led to the adoption of the
term Pharmacogenomics. Pharmacogenomics is among
the first clinical applications of the Human Genome
Project and is certain to have an enormous impact on the
clinical practice of medicine.
Pharmacogenetics:
The study of
how a gene affects the way people respond to medicines, with the
ultimate goal to help tailor medicines to people's unique genetic
make-ups. This will make medicines safer and more effective.
Pharmacogenomics:
The study of genome-derived
data, including human genetic variation, and RNA and protein
expression differences, to predict drug response (e.g.
disposition, safety,
tolerability,
and efficacy) in individual
patients or groups of patients. Thus,
the study of many genes
or entire genomes.
Pharmacogenomics (that combines medicine, pharmacology and
genomics) thus tries to understand the correlation between an
individual patient's genetic make-up (genotype) and their response
to drug treatment. Some drugs work well in some patient populations
and not as well in others. Studying the genetic basis of a response
of a patient to therapeutics allows drug developers to more
effectively design therapeutic treatments.
Thus, through Pharmacogenomics, drugs might one day be
tailor-made for individuals and their conditions, allowing
prescription of the most effective drug dosage and a reduction of
unwanted side effects. Pharmacogenomics is therefore the use of
genetic information to predict drug response. The term drug response
includes two facets: drug effectiveness (efficacy) and drug side
effects. It is estimated that, on average, as much as 40% of the
medicines that individuals take every day are not effective. In
fact, for certain medications, the estimate of non-effectiveness is
well over 50%. A drug simply does not work for every individual and
many people are exposed to the problematic side effects of drugs
while receiving little or no benefit. Pharmacogenomics tries to
identify people whose genetic profiles or "bar codes" predict that
they are inappropriate for a given medication, whether due to poor
efficacy and/or adverse side effects. Pharmacogenomics allows
physicians to prescribe with greater confidence, and pharmaceutical
companies to more effectively target drugs where they will do the
most good. The current one-size-fits-all approach to medicine will
be augmented increasingly by diagnostic analysis that, for many
drugs and many patients, will validate the appropriateness of
certain medications before they are administered. One approach to
pharmacogenomics is to directly study the genetic component of the
problem (the DNA itself) to understand the way in which variations
in DNA sequences contribute to phenotypic traits such as common
diseases and drug responses. Thus,
by applying
the principles of pharmacogenomics, it may be possible to enhance
the productivity of drug discovery and development. Also, by
allowing better identification of genes, pathways, and drug
targets, pharmacogenomics will promote development of the
right drug for the right patient (see Figure 1 below). Pharmacotherapies
to treat psychiatric disorders are incompletely effective. Of the
patients treated with antidepressants, 10–20% react adversely and
25–35% of those who complete an adequate treatment period do not
respond to the medication. This can lead to treatment for patients
being selected in what has been described as a ‘trial and error’
fashion, as physicians or psychiatrists trial different
pharmacological agents with their patients in order to select the
best treatment. The mechanisms that cause non-response in patients
with mood disorders are not well understood but in many cases are
likely to be due to polymorphic genes that affect the way drugs are
metabolised. Variations in drug metabolism can cause prolonged drug
effects, adverse drug reactions, drug toxicity and lack of drug
activation. Such effects can result in patient non-compliance and
poor treatment outcome. Among the promised potential benefits of
genetic research into psychiatric disorders are the pharmacogenetic
and pharmacogenomic approaches to improving the treatment.
Pharmacogenomics
/ pharmacogenetics is aimed at:
-
-
Creating opportunities to increase the value of drugs using genetics
-
- Obtain
greater understanding of disease
-
- predict disease severity, onset, progression
-
- identify genetic subtypes of disease
-
-
aid in discovery of new drug targets
-
-
Distinguish subgroups of patients who respond differently to drug
treatment
-
- Aid in
the interpretation of clinical study results
|