 On the left is the
structure of noradrenaline.
On the left is the
structure of noradrenaline. To find the ligand binding domain of the beta-adrenergic receptor, researchers first examined the structure of noradrenaline, the natural ligand for this receptor.
They examined the ligand structure to make predictions about the type of binding the ligand might make with the receptor.
 On the left is the
structure of noradrenaline.
On the left is the
structure of noradrenaline.
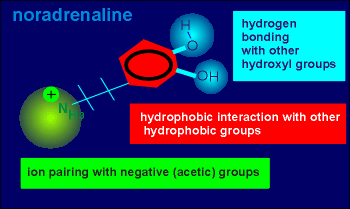
This ligand possesses a positively charged amino group (green), ideal for ion pairing with negative charges in the receptor (i.e. perhaps the negative charges of acetic amino acids such as aspartic acid or glutamic acid).
It also possesses a hydrophobic ring structure (red) ideal for hydrophobic interactions with the ring structure found in the amino acids tyrosine or phenylalanine within the receptor.
Finally, noradrenaline possesses two hydroxyl groups which could potentially hydrogen bond with other hydroxyl groups within the receptor (i.e. to hydroxyl groups of serines or threonines).
The question is, which of the above possibilities are actually used in ligand binding with the receptor.
For an answer, molecular biologists applied an approach discussed earlier to characterize functional domains of the nicotinic receptor, namely site-directed mutagenesis.
Let's first consider ion pairing.
 Within the transmembrane region
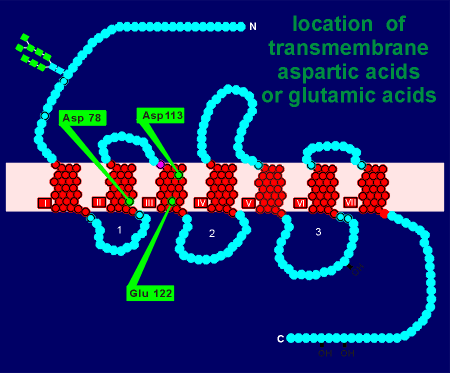
of the beta adrenergic receptor there are only three aspartic acids or glutamic acids
(perhaps not surprising because transmembrane regions are hydrophobic not hydrophilic).
Within the transmembrane region
of the beta adrenergic receptor there are only three aspartic acids or glutamic acids
(perhaps not surprising because transmembrane regions are hydrophobic not hydrophilic).
Using site-directed mutagenesis, receptor cDNA for the beta adrenergic receptor was altered such that the coding for aspartic acid in site 78 was changed to a neutral amino acid.
Likewise, mutated cDNA was prepared where coding for aspartic acid at site 113 was altered (for a neutral amino acid) and, in a third preparation, glutamic acid at site 122 was similarly altered.
The three mutated cDNAs were then used as templates to produce mRNAs.
Each mRNA preparation was then injected into Xenopus oocytes and the expressed (mutated) receptor tested for bind with radioactive noradrenaline.
Alteration of Asp 78 or Glu 122 had little or no effect on ligand binding.
Alteration of Asp 113 drastically reduced noradrenaline binding to the receptor.
Thus, it was concluded that the positive charge in the noradrenaline ion pairs with the negative charge of aspartic acid in site 113 of the receptor.
 For hydrophobic ring interactions a bit more
work was required.
For hydrophobic ring interactions a bit more
work was required.
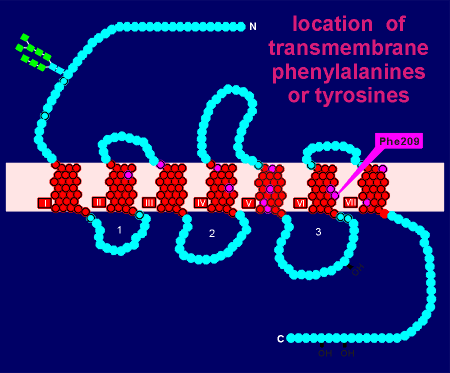
There was a total of 12 phenylalanines or tyrosines (indicated in purple) and thus 12 experiments were required to determine if the substitution of any of these affected ligand binding.
The results were clear... only substitution of phenylalanine at site 209 had a dramatic effect on noradrenaline binding to the expressed receptor.
For hydrogen bonding with serines or threonines even more work was required.
There is a total of 21 serines and threonines in the transmembrane region of the beta adrenergic receptor.
Site-directed mutagenesis showed that serines at positions 204 and 207 in the 5th transmembrane region were involved in binding to noradrenaline.
To make a model of receptor-ligand binding from the above information, it is important to realize that the transmembrane regions of G protein-linked receptors form a ring on the membrane.
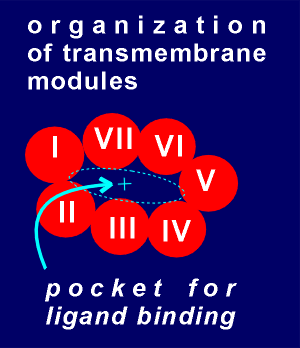 In the diagram to the
right the G protein-linked receptor is viewed from the top such so that each of the seven
transmembrane regions, or modules, are seen in cross section.
In the diagram to the
right the G protein-linked receptor is viewed from the top such so that each of the seven
transmembrane regions, or modules, are seen in cross section.
The transmembrane modules are known to be held in a fairly stable position relative to each other through disulfide cross linking (i.e. cysteines in module 1 forming disulfide bonds with cysteines in modules 2 and 7 etc.).
The idea for ligand binding is that it would take place in the pocket created by the ring-like structure.
Active groups on the side chain of the amino acids would stick into this pocket to bond with the ligand.
In the case of the beta adrenergic receptor we have the acetic group of Asp 113, the hydrophobic ring of Phe 209 and the hydroxyl groups of Ser 204 and 207.
Given to the right is a movie clip showing how noradrenaline might bind
to the receptor in the ligand binding pocket.
Imagine that you are in the ligand binding pocket, looking out to the extracellular space.
You see a noradrenaline molecule in the distance which is homing in on the receptor binding site.
Notice how the noradrenaline makes a perfect fit in the binding site to form (1) an ion pair, (2) two hydrogen bonds and (3) an hydrophobic interaction with the hydrophobic ring of phenylalanine.
The binding of noradrenaline to the receptor will induce conformational changes in the receptor leading to receptor activation.
These types of models have proven very valuable in helping design new receptor agonists and antagonists for G protein linked receptors.